(
PDF version of this article is available)
The Bayes factor test is an interesting thing. Some Bayesians advocate it unequivalently, whereas others reject the notion of
testing altogether, Bayesian or otherwise. This post takes a critical look at the Bayes factor, attempting to tease apart the
ideas to get to the core of what it’s really doing. If you're used to frequentist tests and want to understand Bayes factors, this
post is for you.
The Bayes factor test goes all the way back to Jeffreys’ early book on the Bayesian approach to statistics [Jeffreys, 1939]. Evidence for an
alternative hypothesis against
that of the null hypothesis
is summarized by a quantity known as the Bayes factor. The Bayes factor is just the ratio of the data likelihoods, under
both hypotheses and integrating out any nuisance parameters:
If this ratio is large, we can conclude that there is strong evidence for the alternative hypothesis. In contrast, if the
inverse of this ratio is large, we have evidence supporting the null hypothesis. The definition of what is strong evidence is
subjective, but usually a Bayes factor of 10 or more is considered sufficient.
The standard Bayes factor test against the point-null
Although Bayes factors are sometimes used for testing simple linear regression models against more complex ones, by far
the most common test in practice is the analogue to the frequentist t-test, the Bayes factor t-test. Under the
assumption of normality with unknown variance, it tests a null hypothesis of zero mean against non-zero
mean.
This test is implemented in the BayesFactor R package with the ttestBF method. For the pairwise case it
can be invoked with bf <- ttestBF(x = xdata, y=ydata). For a non-pairwise test just x can be passed
in.
Statistical details of the test
To apply the test we must formally define the two hypotheses in the form of statistical models. Jeffreys’ book is very
pragmatic, and the modelling choices for this test reflect that. Since this is a t-test, we of course assume a normal
distribution for the data for both hypothesis, so it just remains to just define the priors on it’s parameters in both
cases.
We have essentially 3 parameters we have to place priors on: the mean
of the alternative, the variance
under the alternative and
the variance under the null
The under the null
is assumed to be .
We have to be extremely careful about the priors we place on these parameters. Reasonable seeming choices can lead to
divergent integrals or non-sensical results very easily. We discuss this further below.
Variance
Because the variance appears in both the numerator and denominator of the Bayes factor
(the null and the alternative), we can use a “non-informative” improper prior of the form
for both.
This prior causes the marginal likelihood integrals to go to infinity, but this cancels out between the numerator and
denominator giving a finite value.
Mean
The choice of a prior for the mean is a little more subjective. We can’t use an improper prior here, as the mean prior
appears only in the numerator, and without the convenient cancelation with factors in the denominator, the Bayes factor
won’t be well defined. Jeffreys argues for a Cauchy prior, which is just about the most vague prior you can use that still
gives a convergent integral. The scale parameter of the Cauchy is just fixed to the standard deviation in his formulation.
The BayesFactor package uses 0.707 times the standard deviation instead as the default, but this can be
overridden.
What are the downsides compared to a frequentist -test?
The main downside is that the BF test is not as good by some frequentist measures than the tests that are designed to
be as good as possible with respect to those measures. In particular, consider the standard measure of power at
: the probability of
picking up an effect of size
under repeated replications of the experiment. Depending on the effect size and the number of samples
, a Bayesian
-test
often requires 2-4 times as much data to match the power of a frequentist
-test.
Implementation details
The Bayes factor requires computing marginal likelihoods, which is a quite distinct problem from the usual posterior
expectations we compute when performing Bayesian estimation instead of hypothesis testing. The marginal likelihood is
just an integral over the parameter space, which can be computed using numerical integration when the
parameter space is small. For this t-test example, the BayesFactor package uses Gaussian quadrature for
the alternative hypothesis. The null hypothesis doesn’t require any integration since it consists of a single
point.
Jefferys lived in the statistical era of tables and hand computation, so he derived several
formulas that can be used to approximate the Bayes factor. The following formula in terms of the
-statistic
works
well for
in the hundreds or thousands:
The sensitivity to priors
There are two types of priors that appear in the application of Bayes factors; The priors on the hypothesis
,
, and the priors on parameters that
appears in marginal likelihoods, the
. Your prior
belief ratio is
updated to form your posterior beliefs using the simple equation:
The ratio
is not formally considered part of the Bayes factor, but it plays an important role. It encodes your prior
beliefs about the hypotheses. If you are using Bayes factors in a subjective fashion, to update your
beliefs in a hypothesis after seeing data, then it is crucial to properly encode your prior belief in
which hypothesis is more likely to be true. However, in practice everybody always takes this ratio to be
,
following the ideals of objective Bayesian reasoning, where one strives to introduce as little prior knowledge as possible into
the problem. If your writing a scientific paper, it’s reasonable to use 1 as anybody reading the paper can apply there own
ratio as a correction to your Bayes factor, reflecting there own prior beliefs.
The prior ratio of the hypotheses plays an interesting part in the calibration of the Bayes factor
test. One way of testing Bayes factors is to simulate data from the priors, and compare the empirical
Bayes factors over many simulations with the true ratios. This involving sampling a hypothesis,
or
, then sampling
then D. If the
probability of sampling
and
is not equal at that first stage, then the observed Bayes factor will be off by the corresponding
. This is
somewhat of a tautology, as we are basically just numerically verifying the Bayes factor equation as written. Nevertheless, it
shows that if the priors on the hypotheses don’t reflect reality, then the Bayes factor won’t either. Blind application of a
Bayes factor test won’t automatically give you interpretable “beliefs”.
Parameter priors
The dependence of Bayes factors on the parameter priors
and
is really
at the core of understanding Bayes factors. It is a far more concrete problem than the hypothesis likelihoods, as there is no
clear “objective” choices we can make. Additionally, intuitions from Bayesian estimation problems do not carry over to
hypothesis testing.
Consider the simple BF t-test given above. We described the test using the Cauchy prior on the location
parameter of the Gaussian. Often in practice people use large vague Gaussian priors instead when doing
Bayesian estimation (as opposed to hypothesis testing), as they tend to be more numerically stable than the
Cauchy. However, it has been noted by several authors [Berger and Pericchi, 2001] that simply replacing the
Cauchy with a wide Gaussian in the BF t-test causes serious problems. Suppose we use a standard deviation of
in this wide
prior (
large). It turns out that Bayes factor scales asymptotically like:
where is the standard
t-statistic of the data. The large
that was chosen with the intent that it would be ’non-informative’ (i.e. minimally effect the result) turns out to directly
divide the BF!
This is the opposite situation from estimation, where as
increases is behaves more and
more like the improper flat prior .
We can’t use the flat prior in the BF t-test, but if we assume that the sample standard deviation
matches the true standard deviation, we can perform a BF z-test instead. Using a flat prior on
, we get a
BF of:
Because the flat prior appears in the numerator only there is no clear scaling choice for it, so this expression is not in
any sense calibrated (Several approaches to calibration appear in the literature, see Robert [1993]).
It is interesting to see what happens when we apply the Cauchy prior, as used in the standard form of the test. In that
case we see approximate scaling like [Robert et al., 2009]:
A similar formula appears when the data standard deviation is assumed to be known also:
The part can
also be written
in terms of the known standard deviation and the empirical mean, and clearly it is constant asymptotically; the expression
on the whole scales similarly as in the unknown variance case.
Lindley’s paradox
Lindley’s paradox refers to a remarkable difference in the behavior of Bayes factor tests compared
to frequentist tests as the sample size increases. Under a frequentist t-test, we compute the
statistic and compare it against
a threshold that depends on ,
and that decreases as increases,
(Recall the definition of the
statistic: ). In contrast the effective
threshold used by the BF -test
actually increases as increases,
although slowly, following roughly .
This difference is illustrated in the following plot lifted from Rouder et al. [2009]:
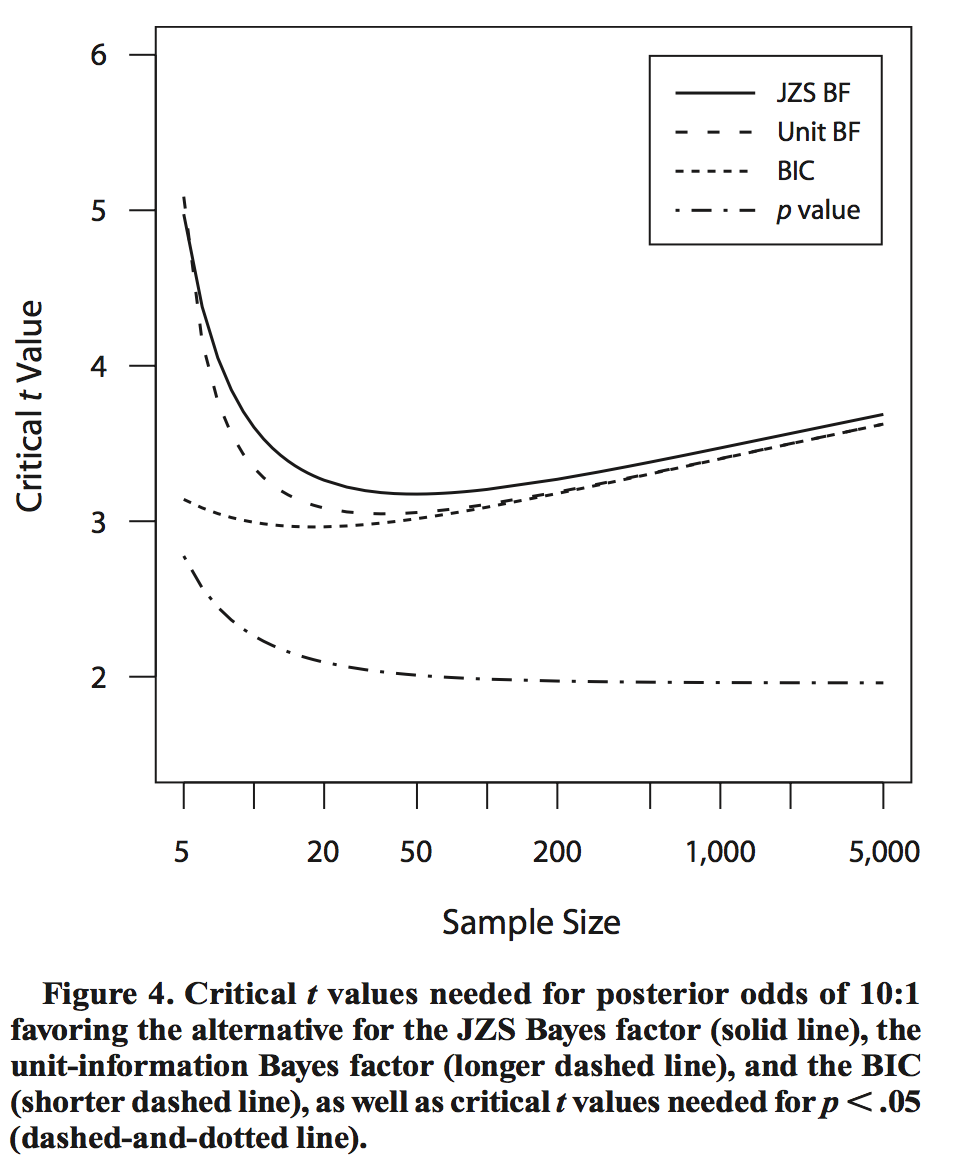
The effect of this difference is that for large sample sizes, a frequentist test can reject the null with small
(say
), while a
BF -test
run on the same data can strongly support the null hypothesis! As a concrete example, if we generate
synthetic data points
with mean , variance
1, then the BF -test
yields 0.028 for the alternative hypothesis (i.e. 36:1 odds for the null being true), whereas a frequentist
-test rejects
the null with
(two-sided test).
It’s all well and good to argue the philosophical differences between Bayesian and frequentist approaches, but when
they disagree so strongly, it becomes a practical issue. The key issue is the handling of small-effects. As
increases and
stays fixed, a
fixed
value of say 2 represents a smaller and smaller measured effect, namely:
Detecting small effects requires a large sample size, and the Bayesian
-test
greatly prefers the null hypothesis over that of a small effect. This is usually described positively as a Occam’s razor effect,
but it does have real consequences when we are attempting to find true small effects. Essentially, the BF test will require
much more data pick up a small effect then a corresponding frequentist test. Often, this is viewed from the opposite side;
that frequentist tests too easily reject the null hypothesis for large sample sizes. This is more of an issue
when testing more complex models such as linear regressions with multiple coefficients. Unless the data was
actually generated from a linear equation with Gaussian noise, large samples will inevitably reject simple
null models. This is in contrast on simple point null hypothesis testing, where we can appeal to asymptotic
normality.
Possible changes from the default Bayes factor test
Here we attempt to answer the following question: are the properties of the default Bayes factor test indicative of BF tests
in general? As we see it, there are two main ways to modify the test: We can use a non-point hypothesis as the null, or we
can change the alternative hypothesis in some fashion. We discuss these possibilities and their combination
separately.
Changing the alternative hypothesis
The default Bayes factor t-test is formulated from a objectivist point of view: it’s designed so that the data
can “speak for its self”, the priors used are essentially as vague as possible while still yielding a usable test.
What about if we choose the prior for the alternative that gives the strongest evidence for the alternative
instead? This obviously violates the likelihood principle as we choose the prior after seeing the data, but it is
nevertheless instructive in that it indicates how large the difference between point NHST tests and BF tests
are.
A reasonable class of priors to consider is those that are non-increasing in terms of
, as they have a
single mode at .
We discuss other priors below. Table 1 shows a comparison of p-values, likelihood ratios from the Neyman-Pearson
likelihood ratio test, and a bound on BFs from any possible prior of this form. It is immediately obvious that according to
this bound, any reasonable choice of prior yields much less evidence for the alternative then that suggested
by the p-value. For example, the difference at p=0.001 is 18 fold, and the difference is larger for smaller
.
|
|
|
|
|
| Method | | | | |
|
|
|
|
|
|
|
|
|
|
| t-statistic | 1.645 | 1.960 | 2.576 | 3.291 |
|
|
|
|
|
| p-value | 0.1 | 0.05 | 0.01 | 0.001 |
|
|
|
|
|
| LR | 0.258:1 | 0.146:1 | 0.036:1 | 0.0044:1 |
|
|
|
|
|
| BF bound | 0.644:1 | 0.409:1 | 0.123:1 | 0.018:1 |
|
|
|
|
|
| |
Although we disregarded multi-modal alternative priors above, they actually form an interesting class of tests. In terms of
conservativeness, their BF is easily seen to be bounded by the likelihood ratio when using a prior concentrated on the maximum
likelihood solution :
This ratio is simplify the Neyman-Pearson likelihood ratio for composite hypothesis,
and it’s values are also shown in Table 1. This values are still more conservative than
-values
by a factor of 2-5. Note that in a Neyman-Pearson test such as the frequentist
-test, this
ratio is not read-off directly as an odds ratio as we are doing here; that’s why this NP odds ratio doesn’t agree directly with
the p-value.
The interesting aspect of multi-modal priors is that they allow you to formulate a test that is more powerful
for finding evidence of the null hypothesis. It can be shown that under the default BF t-test, evidence for
the null accumulates very slowly when the null is true, where as evidence for the alternative accumulates
exponentially fast when the alternative is true. Multi-modal priors can be formulated that fix this imbalance
[Johnson and Rossell, 2010]. One simple case is a normal or Cauchy prior with a notch around the null
set to
have low probability.
If we go a step further and consider the limits of sequences of improper priors, the Bayes factor
can take essentially any value. Some particular “reasonable” sequences actually yield values very similar to
-test’s
p-values [Robert, 1993] as their posterior probabilities, at least between 0.1 and 0.01. Their construction also places 0
probability mass in the neighborhood of the null point in the limit, so it is similar to the multi-modal priors discussed in the
previous paragraph. Although this improper prior does actually avoid Lindley’s paradox, it is more of a theoretical
construction; it is not recommend in practice, even by it’s discoverer [Robert, 1993].
Consequences for Lindley’s paradox
As discussed above, when restricting ourselves to well behaved proper priors, “Lindley’s paradox” as such can not be
rectified by a careful choice of a proper prior on the alternative hypothesis. When one takes a step back and considers the
consequences of this, Lindley’s paradox becomes much clearer, as a statement about objective priors rather than BF tests in
general.
The central idea is this: It requires a lot of evidence to detect small effects when you use vague priors. The idea of running a
long experiment when you use a vague prior is nonsensical from an experimental design point of view: large amounts of data are
useful for picking up small effects, and if you expect such a small effect you should encode this sensibly in your prior. The default
Cauchy prior used in the BF t-test essentially says that you expect a 50% chance that the absolute effect size (Cohen’s
) is larger
than ,
which is a completely implausible in many settings. For instance, if you are trying to optimize conversion rates on a website
with say a 10% base conversion rate, an effect size of “1” is a 4x increase in sales! Some software defaults to 0.707 instead of
1 for the Cauchy scale parameter, but the effect is largely the same
Experimental design considerations are important in the choice of a prior, and the Bayesian justification of the Bayes
factor depends on our priors being reasonable. The choice of using a particular BF test is not a full experimental
design, just as for a frequentist it is not sufficient to just decide on particular test without regard to the
eventual sample size. It is necessary to examine other properties of the test, so determine if it will behave
reasonably in the experimental setting in which we will apply it. So in effect, Lindley’s paradox is just the
statement that a poorly designed Bayesian experiment won’t agree with a less poorly designed frequentist
experiment.
Put another way, if the Bayes factor is comparing two hypothesis, both of which are unlikely under the data, you
won’t get reasonable results. You can’t identify this from examining the Bayes factor, you have to look at
the experiment holistically. The extreme case of this is when neither hypothesis includes the true value of
. In such
cases, the BF will generally not converge to any particular value, and may show strong evidence in either
direction.
An example
It is illustrative to discuss an example in the literature of this problem: a high profile case where the Bayesian
and frequentist results are very different under a default prior, and more reasonable under a subjective
prior. One such case is the study by Bem [2011] on precognition. The subject matter is controversial;
Bem gives results for 9 experiments, in which 8 show evidence for the existence of precognition at the
level! This obviously extraordinary claim lead to extensive examination of the results from the Bayesian
view-point.
In Wagenmakers et al. [2011a], the raw data of Bem [2011] is analyzed using the default Bayesian t-test. They report
Bayes factors towards the alternative (in decreasing order) of 5.88, 1.81, 1.63, 1.05, 0.88, 0.58, 0.47, 0.32, 0.29, 0.13 . So only
one experiment showed reasonable evidence of precognition (5.88:1 for), and if you combine the results you get 19:1 odds
against an effect.
In contrast, Bem et al. [2011] performed a Bayes factor analysis using informative priors on effect sizes, uses values
considered reasonable under non-precognitive circumstances. The 3 largest Bayes factors for the existence of precognition
they got were 10.1, 5.3 and 4.9, indicating substantial evidence!
The interesting thing here is that the vague prior used by Wagenmakers et al. [2011a] placed a large prior
probability on implausibly large effect sizes. Even strong proponents of ESP don’t believe in such large effects,
as they would have been easily detected in other past similar experiments. Somewhat counter-intuitively,
this belief that ESP effects are strong if they exist at all yields a weaker posterior belief in the existence of
ESP!
It should be noted that Wagenmakers et al. [2011b] responded to this criticism, but somewhat unsatisfactory in my
opinion. Of course, even with the use of correct subjective Bayesian priors on the alternative hypothesis, these experiments
are not sufficient to convince a skeptic (such as my self) of the existence of ESP! If one factors in the prior
and
updates their beliefs correctly, you may go from a 1:1-million belief in ESP existing to a 1:76 belief (if you use the naive
combined BF of 13,669 from Bem et al. [2011]).
Most reasonable people would consider the chance that a systematic unintentional error was made during the
experiments to be greater than 1 in a thousand, and so not update their beliefs quite as severely. Regardless, several
pre-registered replications of Bem’s experiments have failed to show any evidence of precognition [Ritchie
et al., 2012].
Recommendations when using point-nulls
The general consensus in the literature is that single Bayes factors should not be reported. Instead the BFs corresponding to
several different possible priors for the alternative hypothesis should be reported. Only if the conclusions are stable for a
wide range of priors can they be reported as definitive. If the results are not stable, then it must be argued why a particular
prior, chosen subjectively, is the best choice.
Bounds giving the largest or smallest possible Bayes factor over reasonable classes of priors, such as the bounds given in
Berger and Jefferys [1991], can also be reported. The consideration of multiple possible priors is generally called “robust”
Bayesian analysis.
Non-point nulls
As we showed above, under essentially any point-null Bayes factor t-test, the results are much more conservative
than frequentist t-tests on the same data. But is this indicative of BF tests in general? It turns out Bayes
factor tests that don’t use point-nulls behave much more like their frequentists counterparts [Casella and
Berger, 1987].
But first, lets just point out that point-null hypotheses are fundamentally weird things. When we start assigning positive
probability mass to single values, we move from the comfortable realm of simple distribution functions we can plot on a
whiteboard, into the Terra pericolosa that is measure theory. As anybody who has done a measure theory course knows,
common sense reasoning can easily go astray even on simple sounding problems. The “surprising” Occam’s razor
effect seen in point-null BF tests is largely attributable to this effect. It can be argued that any prior that
concentrates mass on a single point is not “impartial”, effectively favoring that point disproportionately [Casella and
Berger, 1987].
One-sided tests
The simplest alternative to a point null test is a postive versus negative effect test, where the null is taken to be the entire
negative (or positive) portion of the real line. This test is very similar in effect to a frequentist one sided test. Unlike the
point-null case, both the alternative and null hypotheses spaces are the same dimension; in practice this means we can use
the same unnormalized priors on both without running into issues. This test can be preformed using the BayesFactor R
package with the ttestBF method, althought somewhat indirectly, by taking the ratio of two seperate point null
tests:
bfInterval <
− ttestBF(x = ldata, y=rdata, nullInterval=c(
−Inf,0))
bfNonPoint <
− bfInterval[2] / bfInterval[1]
print(bfNonPoint)
Applying one-sided tests in the frequentist setting is routine (in fact often advocated [Cho and Abe, 2013]), it
is surprising then that their theoretical properties are very different to two-sided tests. If we look at both
Bayesian and frequentist approaches from the outside, using decision theory (with quadratic loss), we see a
fundamental difference. For the one sided case, the Fisher p-value approach and obeys a weak kind of optimality
known as admissibility [Hwang et al., 1992]: no approach strictly dominates it. In contrast, in the two-sided
case neither the frequentist or the standard Bayesian approach is admissible, and neither dominates the
other!
What does this tell us about sequential testing settings?
The use of Bayes factor tests is sometimes advocated in the sequential testing setting, as their interpretation is not
affected by early stopping. “Sequential testing” is when a test of some form of test is applied multiple times during the
experiment, with the experiment potentially stopping early if the test indicates strong evidence of an effect. Using
frequentist tests sequentially is problematic; the false-positive rate is amplified and special considerations
must be made to account for this. See my previous post for details on how to perform the frequentist tests
correctly.
The interpretation of the Bayes factor in contrast is unaffected by early stopping. Naive application of a point-null BF
test does seem to perform reasonable in a sequential setting, as it’s naturally conservative nature results in few false
positives being detected. However, as we pointed out above, one-sided non-point null tests are equally valid, and they are
not particularly conservative. Just applying one sequentially gives you high false-positive rates comparable to the frequentist
test. The false-positive rate is a frequentist property, and as such it is quite unrelated to the Bayesian interpretation of the
posterior distribution.
Indeed, in most sequential A/B testing situations, such as website design tests, the one-sided non-point test seems the
most reasonable, as the two hypothesis correspond more directly to the two courses of action that will be taken after the
test concludes: Switch all customers to design A or design B.
So, should I use a point null hypothesis?
Here we start to move outside the realm of mathematics and start to meander into the realm of philosophy. You have to
consider the purpose of your hypothesis test. Point-nulls enforce a strong Occam’s razor effect, and depending on the
purpose of your test, this may or may not be appropriate.
The examples and motivation used for null hypothesis testing in the original formulation of the problem by Jeffreys
came from physics. In physics problems testing for absolute truth is a much clearer proposition. Either an exotic particle
exists or it doesn’t, there is no in-between. This is far cry from the psychology, marketing and economics problems that are
more commonly dealt with today, where we always expect some sort of effect, although potentially small and possibly in the
opposite direction from what we guessed apriori.
You need a real belief in the possibility of zero effect in order to justify a point null test. This is a real
problem in practice. In contrast, by using non-point null tests you will be in closer agreement with frequentist
results, and you will need less data to make conclusions, so there is a lot of advantages to avoiding point null
tests.
It is interesting to contrast modern statistical practice in high energy physics (HEP) with that of other areas where statistics
is applied. It is common practice to seek “five sigma” of evidence before making a claim of a discovery in HEP fields. In contrast
to the usual
requirements, the difference is vast. Five sigma corresponds to about a 1-in-1million
value. However,
if you view the five sigma approach as a heuristic approximation to the point-null BF test, the results are much more comparable.
The equivalent
statistic needed for rejection of the point null under the Bayesian test scales with the amount of data. HEP experiments often
average over millions of data points, and it is in that multiple-million region where the rejection threshold, when rephrased in
terms of the
statistic, is roughly five.
Concluding guidelines for applying Bayes factor tests
- Don’t use point-null Bayesian tests unless the point null hypothesis is physically plausible.
- Always perform a proper experimental design, including the consideration of frequentist implications of your
Bayesian test.
- Try to use an informative prior if you are using a point null test; non-informative priors are both controversial
and extremely conservative. Ideally show the results under multiple reasonable priors.
- Don’t blindly apply the Bayes factor test in sequential testing situations. You need to use decision theory.
References
Daryl J. Bem. Feeling the future: Experimental evidence for anomalous retroactive influences on cognition and
affect. Journal of Personality and Social Psychology, 2011.
Daryl J. Bem, Jessica Utts, and Wesley O. Johnson. Must psychologists change the way they analyze their
data? a response to wagenmakers, wetzels, borsboom, & van der maas. 2011.
James O. Berger and William H. Jefferys. The application of robust bayesian analysis to hypothesis testing
and occam’s razor. Technical report, Purdue University, 1991.
James O. Berger and Luis R. Pericchi. Objective bayesian methods for model selection: Introduction and
comparison. IMS Lecture Notes - Monograph Series, 2001.
J.O. Berger and R.L. Wolpert. The Likelihood Principle. Institute of Mathematical Statistics. Lecture
notes : monographs series. Institute of Mathematical Statistics, 1988. ISBN 9780940600133. URL
https://books.google.com.au/books?id=7fz8JGLmWbgC.
George Casella and Roger L. Berger. Reconciling bayesian and frequentist evidence in the one-sided testing
problem. Journal of the American Statistical Association, 82(397):106–111, 1987. ISSN 0162-1459. doi:
10.1080/01621459.1987.10478396.
Hyun-Chul Cho and Shuzo Abe. Is two-tailed testing for directional research hypotheses tests legitimate? Journal
of Business Research, 66(9):1261 – 1266, 2013. ISSN 0148-2963. doi: http://dx.doi.org/10.1016/j.jbusres.2012.02.
023. URL http://www.sciencedirect.com/science/article/pii/S0148296312000550. Advancing Research
Methods in Marketing.
Jiunn Tzon Hwang, George Casella, Christian Robert, Martin T. Wells, and Roger H. Farrell. Estimation
of accuracy in testing. Ann. Statist., 20(1):490–509, 03 1992. doi: 10.1214/aos/1176348534. URL
http://dx.doi.org/10.1214/aos/1176348534.
Harold Jeffreys. Theory of Probability. 1939.
V. E. Johnson and D. Rossell. On the use of non-local prior desities in bayesian hypothesis tests. Journal of
the Royal Statistical Society, 2010.
Stuart J. Ritchie, Richard Wiseman, and Christopher C. French. Failing the future: Three unsuccessful
attempts to replicate bem’s ‘retroactive facilitation of recall’ effect. PLoS ONE, 2012.
Christian P. Robert. A note on jeffreys-lindley paradox. Statistica Sinica, 1993.
Christian P. Robert, Nicolas Chopin, and Judith Rousseau. Harold jeffreys’s theory of probability revisited.
Statistical Science, 2009.
Jeffrey N. Rouder, Paul l. Speckman, Dongchu Sun, Richard D. Morey, and Geoffrey Iverson. Bayesian t tests
for accepting and rejecting the null hypothesis. Psychonomic Bulletin & Review, 2009.
Eric Jan Wagenmakers, Ruud Wetzels, Denny Borsboom, and Han van der Maas. Why psychologists must
change the way they analyze their data: The case of psi. Journal of Personality and Social Psychology, 2011a.
Eric Jan Wagenmakers, Ruud Wetzels, Denny Borsboom, and Han van der Maas. Why psychologists must
change the way they analyze their data: The case of psi: Clarifications for bem, utts, and johnson (2011). Journal
of Personality and Social Psychology, 2011b.